Abstract
Background
Recent reports have highlighted an adverse impact of TP53 mutations on the prognosis of patients with myeloid malignancies. TP53 mutational analysis is useful in classifying these patients with respect to treatment strategies. High Resolution Melt (HRM) analysis is based on differences in the patterns of melting curves obtained when the targeted double strand DNA (dsDNA) dissociates during heating. HRM analysis is more cost- and time-effective than next-generation sequencing (NGS). In this study, we used HRM analysis to evaluate the impact of TP53 mutations on the clinical features and prognosis of patients at our institute with myeloid malignancies.
Methods
In this retrospective case study, HRM analysis was performed on 23 patients with MDS and 131 with AML, from October 2011 to January 2018. We targeted ten genes recurrently mutated in myeloid malignancies (TP53, NPM1, FLT3-ITD, IDH1, IDH2, NRAS, KRAS, WT1, DNMT3A, CEBPA) using HRM analysis for screening. For TP53 mutation detection, the entire coding region of TP53 gene from exons 1 to 11 was analyzed using the HRM method. Positive samples were validated using Sanger sequencing. We excluded frequently reported single nucleotide polymorphisms (P47S and R72P), silent mutations, and mutations which were not reported in the Catalogue of Somatic Mutations in Cancer database. NGS of TP53 mutations was done in 18 cases to validate mutations found by HRM analysis. Other non-TP53 gene mutations were similarly analyzed. The primary clinical endpoint of this study was overall survival (OS), measured from the time of diagnosis till time of death due to any cause, or till the last follow-up. The secondary clinical endpoint was relapse-free survival (RFS), measured from time of complete remission to relapse, or the last follow-up. The OS and RFS were compared using the log-rank test.
Results
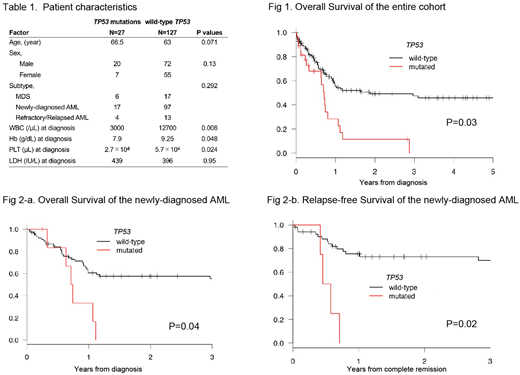
We evaluated gene mutations in 131 AML and 23 cases of MDS using HRM analysis. Costs for one HRM analysis were $25 (USD, excluding labor) and required approximately 7 hours processing time per sample. If the screen was positive, add-on cost was an additional $20 with 2 hours of processing time. In total, we identified 28 TP53 somatic mutations in 27 cases from this cohort using HRM analysis (17 newly-diagnosed AML, 4 refractory/relapsed AML, and 6 MDS). Out of the total 154 cases, 18 were also analyzed by NGS; TP53 mutations were detected in 4 cases (median variant allele frequencies = 0.42) and no TP53 mutations were detected in the remaining 14 cases, consistent with the results of HRM analysis. Among the 28 somatic TP53 mutations, 26 were missense, 1 frameshift, and 1 affecting splice sites. Most missense mutations detected (22/26) were localized to the DNA-binding domain of the gene. Cases with TP53 mutations frequently had a complex karyotype (P=0.001), but rarely had NPM1 mutations (P=0.039). In comparison with patients with myeloid malignancies but no TP53 mutations, patients with TP53 mutations had lower counts of white blood cells (3,000 ·μL-1 vs 12,700 ·μL-1 ; P=0.006), lower hemoglobin (7.9 g·dL-1 vs 9.3 g·dL-1 ; P=0.048), and lower platelet counts (2.7×104 ·μL-1 vs 5.7×104 ·μL-1 ; P=0.024). TP53 mutations were associated with shorter OS (median OS, 260 days vs 687 days; P=0.02). In newly-diagnosed AML patients, TP53 mutations were associated with lower counts of both white blood cells (3,200 ·μL-1 vs 26,550 ·μL-1; P=0.006) and lower platelet counts (2.0×104 ·μL-1 vs 6.2×104 ·μL-1; P=0.008) in the peripheral blood. Analysis of bone marrow aspirate revealed that more cases with TP53 mutations had a low myeloid:erythroid ratio than those with TP53 wildtype (53.8% vs 12.7%; P=0.02), suggesting that cases with TP53 mutations have a higher proportion of erythroid precursors, such as in acute erythroid leukemia or erythroid/myeloid leukemia (M6) , as per the FrenchAmerican-British (FAB) classification. Among the patients treated with intensive therapy (either conventional chemotherapy or allogeneic stem cell transplantation), cases with TP53 mutations had shorter overall survival (median OS, 265 days vs not reached; P= 0.04) and shorter relapse-free survival (median RFS, 188 days vs not reached; P=0.02).
Conclusion
Patients with TP53 mutations have a much poorer prognosis than those without, and exhibit refractoriness to conventional therapies. HRM analysis is a cost- and time-effective method to detect TP53 mutations.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal